Baffin Bay has long been considered the ‘Jewel of the Texas Coast,’ bringing in tourists and locals alike for recreational activities such as fishing, boating, hiking, hunting, kayaking and birdwatching along the gorgeous south Texas shoreline. However, due to anthropogenic pollution throughout the watershed, water quality in Baffin Bay has experienced a recent and rapid decline. It has experienced harmful algal blooms called brown tides, hypoxia, and fish kills. Three of its major tributaries (San Fernando, Los Olmos, and Petronila Creeks) are listed as impaired due to high concentrations of fecal indicator bacteria. Our lab has the privilege of being a part of the Bringing Baffin Back initiative and our research from the following three projects will go to towards a watershed protection plan for restoring the bay.
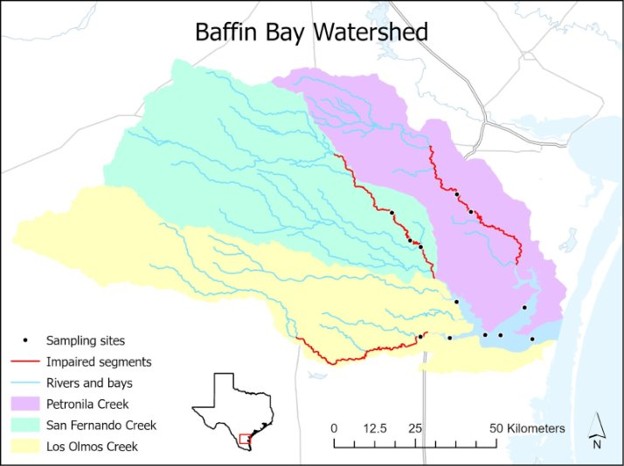
Map of Baffin Bay Watershed 
Sample collection 
Nora collecting Nilgai poop 
Nora presenting at National ASM 2025 
San Fernando Creek 
Petronilla Creek 
Baffin Bay 
Water sample bottles 
Los Olmos bridge 
Nora and Bella on a sample collection trip
Who Pooped in Baffin Bay? Microbial Source Tracking reveals the sources and drivers of fecal pollution throughout the watershed
Fecal pollution is remains a steadily growing problem along the Texas Coastline. Recreational waters and beaches impaired by fecal pollution have a higher risk of illness for people engaging in water activities. Polluted water can harbor germs that cause ear, eye, skin, and gastrointestinal illnesses.
Monitoring and managing fecal pollution is difficult in an area like Baffin Bay, where multiple sources of fecal pollution accumulating in the water, also known as nonpoint source pollution, is the main cause of degradation. We decided to use a method called microbial source tracking (MST) to determine the contribution of different sources of fecal pollution that may be contributing to the decline in water quality throughout the bay. MST uses host-associated genetic markers to quantify fecal pollution from each source.
To do this, we had to examine the probably sources of fecal pollution based on land use. The land surrounding the watershed is mostly rural and has about 11,000 aging septic systems. Expanses of shorebird habitat provide homes to wild birds such as gulls, herons, and egrets. The watershed also has extensive pasture for cattle and populations of invasive species such as feral hogs and nilgai. Therefore, we chose human (HF183), gull (LeeSeaGull), cow/ruminant (BacCow), and pig (Pig-2-Bac) markers to quantify fecal pollution using droplet digital PCR.
For eighteen months, we collected water samples from twelve sites located in Baffin Bay and its tributaries: San Fernando, Petronila and Los Olmos Creeks. This gave us a grand total of 216 water samples to examine for fecal pollution!
Following this, we used the data to preform a quantititave microbial risk assessment (QMRA) to determine the risk of getting a gastrointestinal illness from using these waters.
Using methods like MST and QMRA together can help use understand the big picture in an ecosystem where water quality is complex and influenced by a multitude of variables.
Our graduate student, Nora has completed processing and presented our findings at the American Society for Microbiology’s national meeting. The results are in and a manuscript is in progress! Stay tuned for published results!
Using the Class-1 integron-integrase Gene intI1 to bridge the gap between fecal pollution and antimicrobial resistance gene pollution in Baffin Bay
Tightly linked to fecal pollution, antimicrobial resistance in the environment is another steadily growing form of pollution. Antimicrobial resistance occurs when a microbe is no longer susceptible to an antimicrobial compound such as an antibiotic, a heavy metal like silver or lead, or a disinfectant like bleach or isopropyl alcohol. When a microbe acquires the ability to resist and antimicrobial, the effectiveness of the antimicrobial is reduced.
Antibiotic resistant infections are listed as a threat to human health by all major global health organizations. Despite this, there is no established method for routinely monitoring antimicrobial resistance in the environment.
This is where the class-1 integron-integrase gene may provide (intI1) may provide a solution. The intI1 gene originated in a clinical environment, but established itself into the natural environment like Baffin Bay. Its job is to help facilitate a process called the Horizontal Gene Transfer or HGT. The HGT is a natural process that allows microbes to share genetic information with each other and it plays a crucial role in the spread of antimicrobial resistance.
Previous studies have suggested that intI1 makes a good proxy for antimicrobial and anthropogenic pollution, so we decided to try it using samples from the Baffin Bay Study!
Our intern, Kristen, used a droplet digital PCR assay to quantify the concentration of intI1 and compared it to the previous fecal pollution study using human, cow, gull and pig markers. Nora and Kristen have completed the statistical analysis and the manuscript has been started. Stay tuned for more exciting updates on this project!
Resistome is not futile! Using metagenomics to characterize antimicrobial resistance genes in Baffin Bay
Like any good scientists, we collected duplicates of our Baffin Bay samples, just in case. But what to do if nothing goes wrong in an experiment? Use the extra samples for another experiment of course! This project uses a subset of 144 water samples collected from the microbial source tracking project above to evaluate a growing, global threat: antimicrobial resistance.
Antimicrobial resistance threatens global health, as it prevents antibiotics and antiseptics from effectively killing germs or slowing their growth. The ability for microbes to acquire antimicrobial resistance is encoded in their DNA. These are called antimicrobial resistance genes or ARGs. They can spread through microbial communities and are recognized as a new kind of environmental pollutant.
This experiment uses the metagenome, or all of the genetic material in a sample, to characterize the resistome. The resistome is the collection of all the ARGs in a sample. Resistome diversity can vary greatly depending on the environment and the stressors on the microbial community.
The DNA from these samples was extracted and is currently having the metagenome sequenced at an outside laboratory. Results from the integrase study show that there is a high potential for antimicrobial resistance genes to be present in the Baffin Bay watershed. When these results are in, Nora will use this data to understand how ARGs interact within the watershed.
These results will contribute to the global dataset of antimicrobial resistance genes in the environment and assess the depth of the issue in a vulnerable south Texas ecosystem.
