By now, you’ve probably heard that antibiotic resistant bacteria (ARB) are a global human health concern. They’ve been found in high numbers in hospitals and schools and a lot of work has been done to investigate their presence in these built environments. It turns out ARB can also be found in natural environments, such as lakes, rivers, and coastal bays. The main way we contribute to their spread in these environments is through… our poop. Wastewater that flows into aquatic environments carries both ARB (which can transfer their resistance to other non-antibiotic resistant bacteria) and residual antibiotic compounds we’ve taken (low levels of exposure to antibiotics selects for the survival and spread of ARB).
So, why do we care about ARB being in a natural environment? In a system such as a coastal bay, ARB can cause seafood- and recreation-related human infections that are essentially untreatable. Important wild and aquacultured fishery species (such as sportfish, shrimp, and other tasty invertebrates) are also harmed by ARB, which can increase their mortality.
To help tackle the spread of ARB, we are using next-generation metagenomic DNA sequencing and bioinformatics to detect ARB, specifically, antibiotic resistance (AR) genes, in three Texas coastal bays: Galveston Bay, Copano Bay, and Nueces Bay (Packery Channel, which receives relatively little freshwater, is our control site). Each bay receives a different major type of wastewater inflow based on its surrounding human development. Freshwater inflows to these bays are largely urban (Galveston Bay), agricultural (Copano Bay), and industrial (Nueces Bay). Each type of inflow carries unique compositions of chemical inducers of AR.
We’ve sampled oysters (an ideal sentinel species since they concentrate contaminants via suspension-feeding) and ambient water from each bay, extracted total bacterial metagenomic DNA from each sample, and sent the DNA out for sequencing. I’m currently analyzing the data and will complete my analyses and the project itself in the coming weeks. Until then, here is a figure detailing the overall abundance of AR genes in each sample type per site as well as the list of antibiotic drugs AR genes in the samples conferred resistance against.
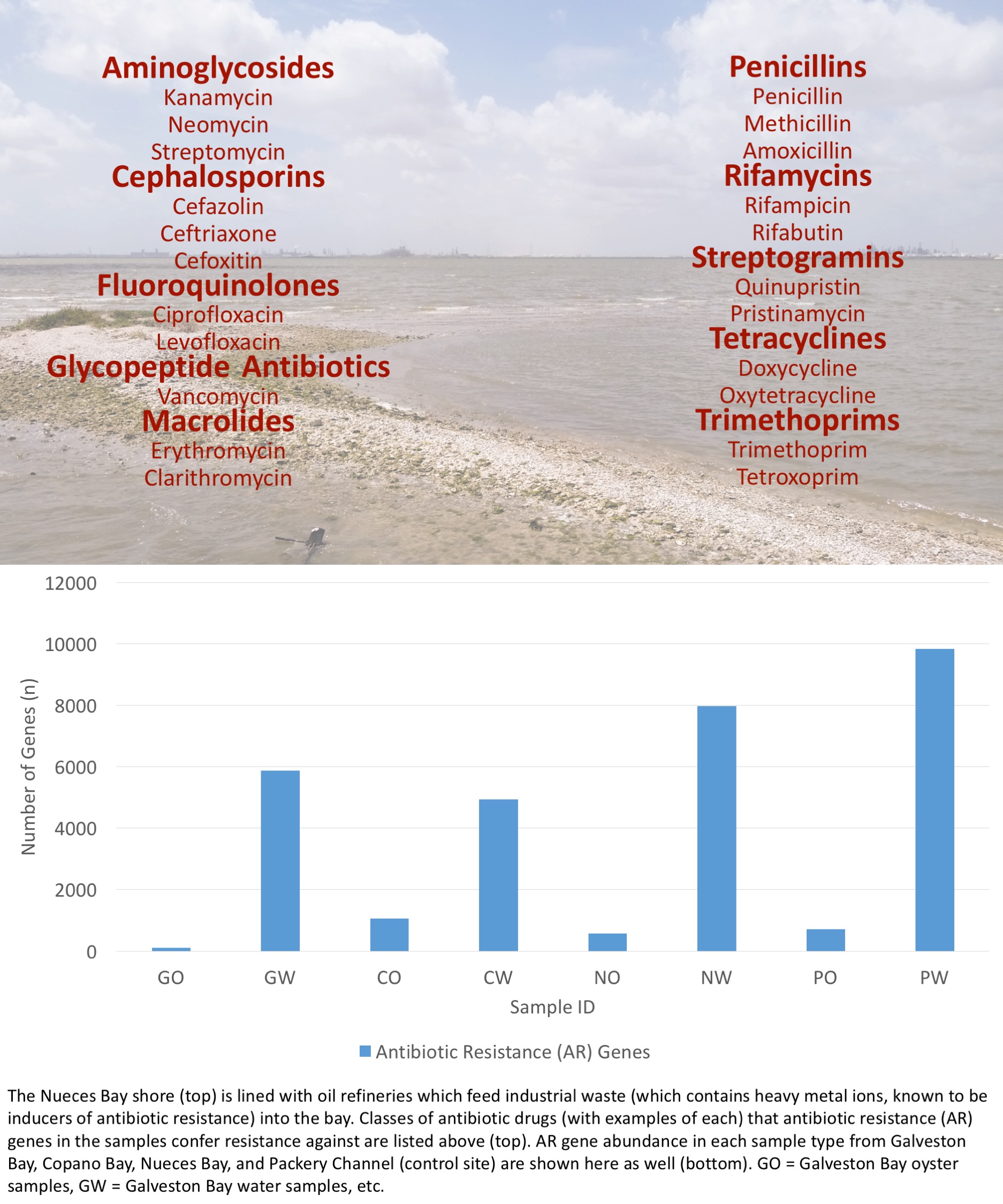
We are extremely excited by what we’re seeing in the samples so far and we can’t wait to uncover more.
I’ll keep you posted,
James

